News
Temporarily no donations to SiDock@home in cryptocurrency
Dear participants,
we temporarily stop accepting donations to SiDock@home in cryptocurrency. If you have automated donations to SiDock@home in cryptocurrency, please disable them. Our previous GRC/BTC/ETH addresses will be unavailable.
With best wishes,
Team of SiDock@home
9 Oct 2025, 9:25:07 UTC
· Discuss
Target # 23: Ebola GP1
Dear participants,
as target # 22 is almost finished, we are glad to introduce the next target. Most of you have voted for Ebolavirus glycoprotein (GP) (174 out of 425 votes, wow!)
The Ebola virus is a highly virulent pathogen responsible for causing Ebola hemorrhagic fever, a severe and often fatal disease. A key factor in the virus's ability to infect host cells and cause disease is its surface glycoprotein (GP), making it an attractive target for antiviral drug development. The Ebola GP is a trimeric protein composed of two subunits per monomer: GP1, responsible for receptor binding, and GP2, which mediates fusion between the viral and host cell membranes. Initially synthesized as a precursor protein, the GP is cleaved by host proteases (furin, cathepsin) into its functional subunits, a process essential for its role in mediating viral entry. The GP facilitates the virus's attachment to the host cell surface, followed by conformational changes that enable membrane fusion, allowing the virus to enter the host cell (to the host endosomal Niemann-Pick C1 (NPC1) receptor or via direct membrane binding; Vaknin et al.; ACS Infect. Dis. 2024, 10, 5, 1590–1601).
Targeting the GP for drug development is advantageous due to its essential role in viral infection, its highly conserved structure among different Ebola virus strains, and the availability of specific binding cavities that can accommodate small-molecule inhibitors. Structural studies using techniques such as X-ray crystallography have identified these binding cavities and elucidated the GP's conformation in both its free and inhibited states. These insights enable the design of drugs that can specifically bind to and inhibit the GP by stabilizing it in its pre-fusion conformation or interfering with its cleavage, thereby preventing the necessary conformational changes for membrane fusion. We will employ high-resolution structures to conduct virtual screening experiments coupled to molecular dynamics simulations to ultimately identify potential GP inhibitors/modulators.
Promising compounds identified through these computational methods will hopefully undergo further validation using biochemical assays, pseudovirus entry assays, and structural analyses to confirm their inhibitory activity. Targeting the GP offers specificity, as it minimizes off-target effects on host cells and reduces the likelihood of resistance development. Moreover, due to the conserved nature of the GP, drugs targeting it could be effective against multiple Ebola virus strains and variants.
We hope that our computations will contribute to the fight against Ebola!
11 Jul 2024, 9:02:55 UTC
· Discuss
Vote for the next target!
Dear all, we have obtained results for different targets of SARS-CoV-2. The computations continue, and we want to ask your opinion on the next target. You can vote for one of them until February 5th, 2024.
- SARS-CoV-2 main protease (3CLpro) further studies
This is a crucial therapeutic target against SARS-CoV-2. 3CLpro (cysteine protease; EC 3.4.22.69) in particular is crucial for the cleavage of coronavirus polyproteins to form mature non-structural proteins that are themselves essential for viral replication mechanisms. We still need much more research on this target towards new more potent inhibitors.
- Ebolavirus glycoprotein (GP)
Ebola virus is a dangerous pathogen to humans and this target could be a perfect study case for PPI-type drug design scenario. The EBOV glycoprotein (GP) is the only virally expressed protein on the virion surface and is critical for attachment to host cells and catalysis of membrane fusion.
- Swine acute diarrhea syndrome coronavirus (SADS-CoV PLpro)
This target would enable us to study the design on multiple related viral targets. Swine acute diarrhea syndrome coronavirus (SADS-CoV), a newly emerging enteric coronavirus, is considered to be associated with swine acute diarrhea syndrome (SADS) which has caused significantly economic losses to the porcine industry. The pathogen indicates towards host-jump potential.
26 Jan 2024, 12:36:20 UTC
· Discuss
Project server maintenance 17th January 2024
Dear participants, tomorrow, 2024.01.17 due to server maintenance, the project will stopped for several hours.
Thank you for attention and project participation!
16 Jan 2024, 12:54:07 UTC
· Discuss
Project status: December 2023
Dear SiDock@home participants,
We've successfully achieved 21 milestones in our ongoing drug discovery initiative, and this strong, open, and community-supported drug discovery project is going on. Our research has been routine lately: virtual screening on the same library for a pleiade of corona-related targets. These efforts are however crucial to our research progression. Currently, we're in the process of drafting publications for two of our completed objectives (3CLpro and PLpro) and are setting the stage for upcoming drug targets (here we are also planning a pool where You the participants, will help us decide on the upcoming target work).
Last but not least, we thank everyone who has donated cryptocurrency or money. The donations sum up to 639 Euro and 12,140 Gridcoin now. We plan to use them for purchase of compounds and in-vitro screening. As always, we are grateful to all of you for your computational contributions and discussions!
We look forward to the future work on SiDock@home.
Merry Christmas to all and all the best to All!
With best wishes,
Natalia, Marko, Črtomir and hoarfrost
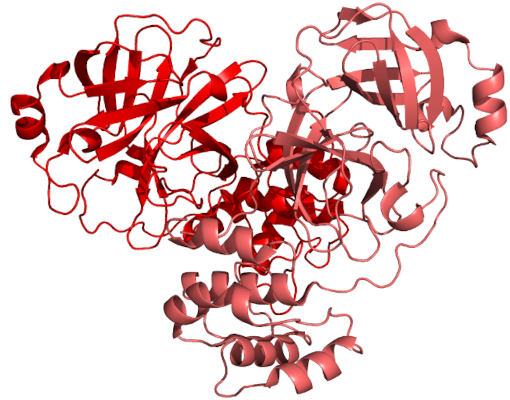
26 Dec 2023, 19:37:17 UTC
· Discuss
... more
News is available as an RSS feed
What is SiDock@home?
Sidock@home is an international volunteer computing project aimed at drug discovery. The first mission of the project is discovery of possible drugs against the SARS-CoV-2 virus. It started as an extension of the project ![]() COVID.SI to engage the BOINC community into the drug discovery.
COVID.SI to engage the BOINC community into the drug discovery.
You can help with your computer. With the help of BOINC, you will download a subset of compounds on your computer, examine the compounds in the context of the studied target and send the results to a server where they are collected for later analysis.
For account creation please use a Crunch_4Science invitation code. It is not needed when registering by BOINC Manager.
Already joined? Log in.
User of the Day
Donations to SiDock@home
We are going to use donation money for biological evaluation of results.
©2026 SiDock@home Team

